Adrian Wolny
SEISMO: Increasing Sample Efficiency in Molecular Optimization with a Trajectory-Aware LLM Agent
Jan 31, 2026Abstract:Optimizing the structure of molecules to achieve desired properties is a central bottleneck across the chemical sciences, particularly in the pharmaceutical industry where it underlies the discovery of new drugs. Since molecular property evaluation often relies on costly and rate-limited oracles, such as experimental assays, molecular optimization must be highly sample-efficient. To address this, we introduce SEISMO, an LLM agent that performs strictly online, inference-time molecular optimization, updating after every oracle call without the need for population-based or batched learning. SEISMO conditions each proposal on the full optimization trajectory, combining natural-language task descriptions with scalar scores and, when available, structured explanatory feedback. Across the Practical Molecular Optimization benchmark of 23 tasks, SEISMO achieves a 2-3 times higher area under the optimisation curve than prior methods, often reaching near-maximal task scores within 50 oracle calls. Our additional medicinal-chemistry tasks show that providing explanatory feedback further improves efficiency, demonstrating that leveraging domain knowledge and structured information is key to sample-efficient molecular optimization.
Sparse Object-level Supervision for Instance Segmentation with Pixel Embeddings
Mar 26, 2021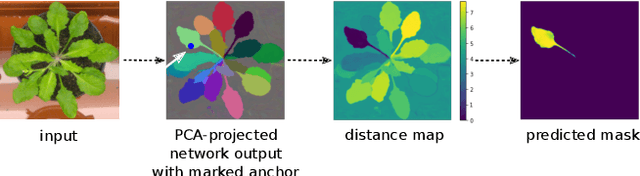
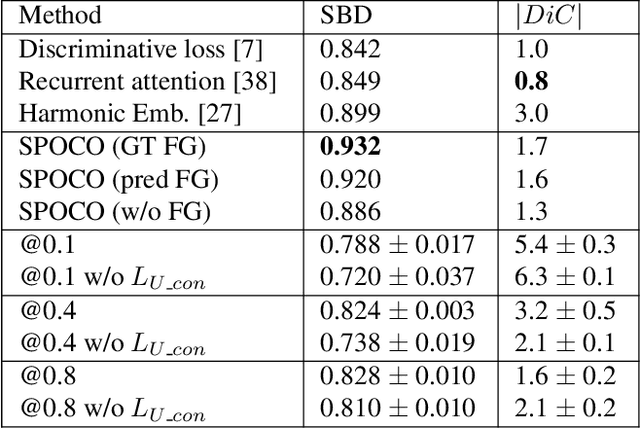
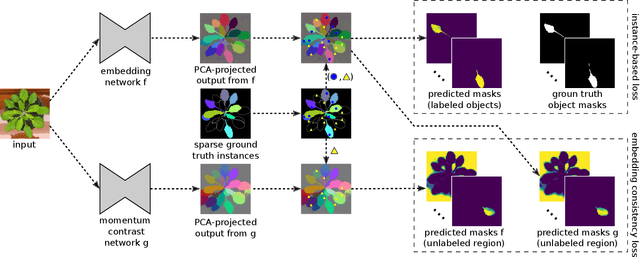

Abstract:Most state-of-the-art instance segmentation methods have to be trained on densely annotated images. While difficult in general, this requirement is especially daunting for biomedical images, where domain expertise is often required for annotation. We propose to address the dense annotation bottleneck by introducing a proposal-free segmentation approach based on non-spatial embeddings, which exploits the structure of the learned embedding space to extract individual instances in a differentiable way. The segmentation loss can then be applied directly on the instances and the overall method can be trained on ground truth images where only a few objects are annotated, from scratch or in a semi-supervised transfer learning setting. In addition to the segmentation loss, our setup allows to apply self-supervised consistency losses on the unlabeled parts of the training data. We evaluate the proposed method on challenging 2D and 3D segmentation problems in different microscopy modalities as well as on the popular CVPPP instance segmentation benchmark where we achieve state-of-the-art results. The code is available at: https://github.com/kreshuklab/spoco
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge